Reconstructing the Pancreatic Cancer Architecture 3
Reconstructing the Pancreatic Cancer Architecture 3
Carina Shiau, Jingyi Cao, Mark Gregory, Sean Kim, Youngmi Kim, Jason Reeves, Eric Miller, Jennifer Su, Jimmy A. Guo, Steven Wang, Peter Wang, Nicole Lester, Tyler Jacks, David T. Ting, Martin Hemberg, William L. Hwang
Koch Institute at MIT, Massachusetts General Hospital
In collaboration with Nanostring Technologies, we are optimizing a single-cell spatial transcriptomics method (spatial molecular imaging) for pancreatic cancer specimens to develop a high-resolution understanding of how the tumor microenvironment is remodeled by treatment and identify candidate ligand-receptor interactions that mediate therapeutic resistance.
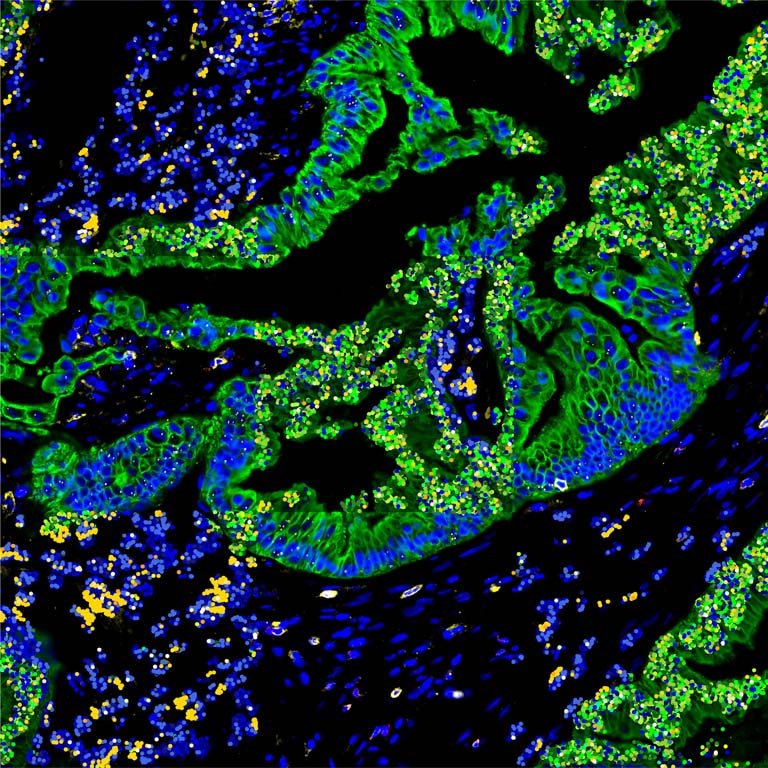
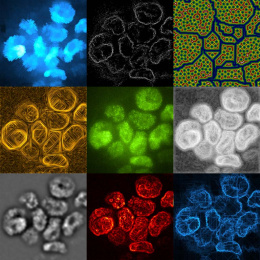
We performed high-resolution, multi-modality imaging of proteins (immunofluorescence) and messenger RNA transcripts (spatial transcriptomics) in a slice of human pancreatic cancer with each modality displayed in alternating squares. For protein squares, cancer cells are stained in green, immune cells in yellow/red, and other cell types in the tumor microenvironment are blue. For messenger RNA squares, a subset of RNA transcripts (out of approximately 1000 different genes detected) that are localized with micron precision are displayed as colored dots. Canonical marker genes for cancer cells are shown in shades of green and immune cells are shown in shades of yellow. Housekeeping genes are displayed in shades of blue. The fact that the messenger RNA squares largely maintain and recapitulate the architecture of the tumor microenvironment in a cell-type specific manner as defined by the protein squares suggests that the spatial transcriptomics method has high specificity.