Mapping Transcription Activity in a Metastatic Tumor
Mapping Transcription Activity in a Metastatic Tumor
Collections: Metastasis
Liangliang Hao, Nour-Saïda Harzallah, and Carmen Martin-Alonso (Bhatia lab); Jonathan Braverman (Yilmaz lab), George Eng (Yilmaz lab), Kathy Cormier (histology core), Jeffrey Kuhn (microscopy core), Vincent Butty (BMC core)
Koch Institute at MIT
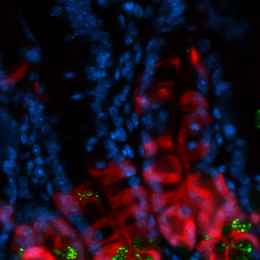
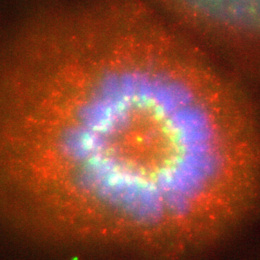
This colorectal cancer liver metastasis tissue histological section is overlayed with its corresponding transcriptional information: We map the activity of disease-causing genes to the genes’ location on the underlying tumor-bearing liver tissue histology. Each dot represents approximately ~10 cells and is colored according to a gradient of gene expression levels (highest in red and lowest purple).
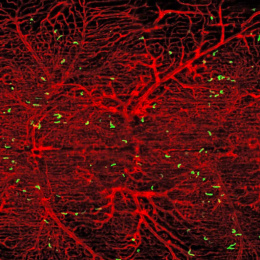
Our overarching research goal is to leverage locally expressed, dysregulated protease activity to develop precision diagnostics and therapeutics for cancer metastasis. However, due to a lack of methods to measure and localize protease activity directly within the tumor microenvironment, the design of protease-activated agents has been strictly empirical, yielding suboptimal results when translated to patients. To address the need for precisely resolved protease activity profiling in cancer, we apply the emerging spatial transcriptomics and multi-color immunofluorescent tools to assess dysregulated protease activity in the context of CRC liver metastasis. Ultimately, we will leverage this knowledge to activate therapeutics, ranging from imaging agents that monitor disease margins to immunotherapies with improved specificity.