Capturing the Genome Folding Process 2
Capturing the Genome Folding Process 2
Simon Grosse-Holz, Laura Caccianini
Mirny Laboratory
Institute of Medical Engineering and Science, MIT Department of Physics
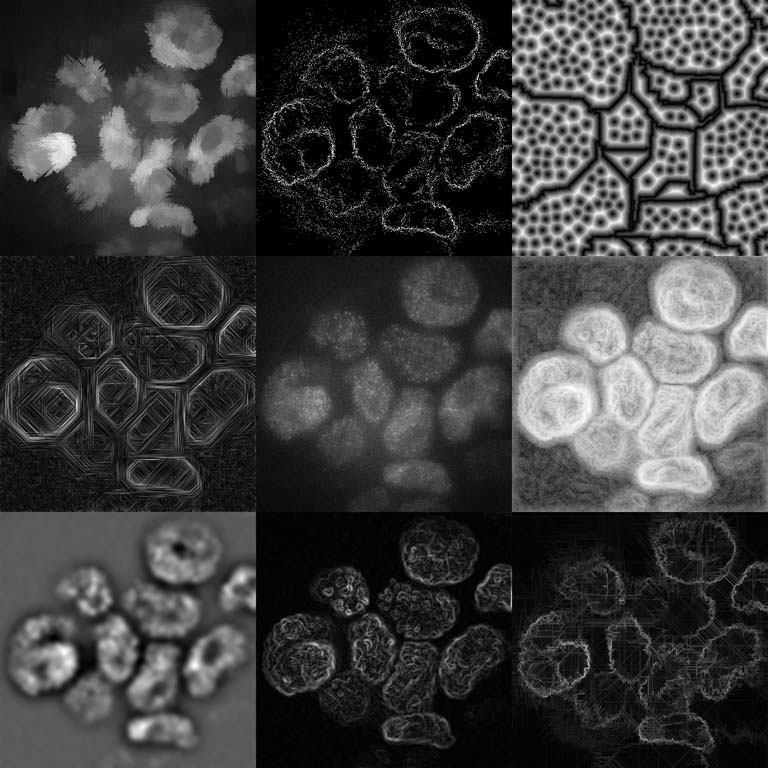
This picture is a collage of different image processing approaches aimed at understanding how the genome is folded in 3D inside a cellular nucleus.
The center panel of the image shows a still frame from a movie spanning about a minute. One can clearly identify some bright areas, which are cellular nuclei. The tiny bright spots inside the nuclei are individual cohesin complexes, which we track over time. In order to identify the whole nuclei and their movement in the movies, we applied a variety of different image processing techniques to the original data. The outer panels show some of these filters applied to the real data image in the center.
These images are essentially an impression of what the computer “sees” when primed to pay attention to different types of detail: some methods are designed to find and trace the edges of the nuclei, while others focus more on identifying them as bright areas in the image, and yet others take completely different approaches to the problem, like simply finding identifiable (and therefore trackable) feature points.
Noting that most of the processed images are missing or misidentifying some of the nuclei, the ultimate question this assembly raises is: What structural features let us, as humans, identify and count the 13 nuclei that are clearly visible in the raw data?